How PredRet works
Stay tuned for more. Please refer to the open access paper for now.
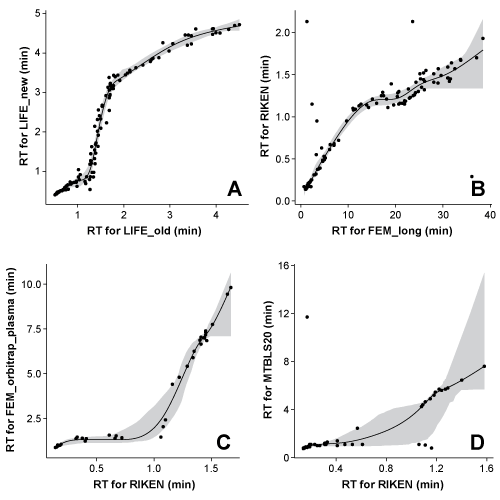
The RTs (experimentally determined in your system and in another users systems) of a number of compounds are used to create projection models between the RTs in the two systems. You can see examples of such models in the plot to the right (Figure 1). These models can then be used to predict the RT of a compound if the RT is known in the other system, but not in yours. Building these models between all chromatographic systems in the database thus allows us to predict the RT of a high number of compounds in your system.
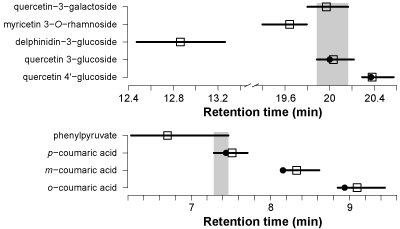
Below you can see an example of predicted RTs for five isomers (Figure 2). Delphinidin-3-glucoside can be clearly distinguished from the others based on the predicted RT and prediction intervals. Myricetin 3−O−rhamnoside and quercetin 4′-glucoside can in theory be distinguished from the others while quercetin 3-glucoside and quercetin 3-galacoside are not separated.